St. Jude Cloud visualizations empower scientist’s work
St. Jude Cloud enables investigators to create beautiful scientific visualizations from rich multi-omics datasets.
Scientific experiments increasingly generate large swaths of data. When publishing results, researchers share a written manuscript alongside figures that visually represent and explain their findings. Having clear and aesthetically pleasing data visualizations is critical for scientists to communicate their findings so that others in the field can understand and appreciate the significance of the work.
The era of “Big Data” has made this process complex because representing enormous datasets is difficult due to the sheer amount of information to convey. It is also a challenge because each step in the process has complexities, such as processing the raw data, that make it difficult. These barriers loom large over small labs with a lack of access to funds to perform expensive large-scale sequencing experiments or pay for downstream data storage and processing power.
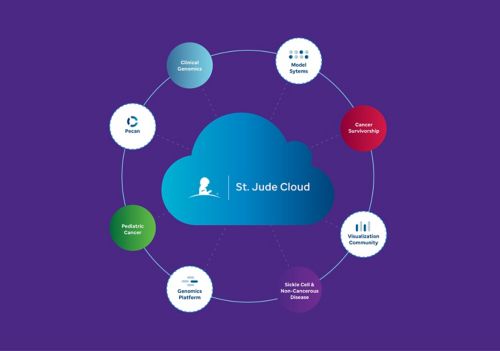
St. Jude devised a solution to store and share raw data, enabling investigators to create beautiful scientific visualizations within St. Jude Cloud. St. Jude Cloud contains multi-omics data from patients with different cancer subtypes and other catastrophic childhood diseases. St. Jude sequenced the patients’ tumors and germline genetics, connected to clinical outcome data for some samples.
To ensure that data in St. Jude Cloud is useful, the team made intuitive interfaces that researchers can explore using the platform and then convert the data to publication-quality visualizations. St. Jude Cloud has already helped scientists publish high-impact papers.
One such scientist is Asa Karlstrom, PhD, St. Jude Department of Developmental Neurobiology, a research program manager who has extensively used St. Jude Cloud’s data analysis visualization tools.
“One example is my supervisor’s, Dr. Michael Dyer’s, Nature Communications paper, which used the Cloud visualizations and made all of its data publicly available,” said Karlstrom. Dyer is the co-leader of Childhood Solid Tumor Network (CSTN) and the St. Jude Department of Developmental Neurobiology chair. “Anyone who goes into the visualization applications can look for the underlying data, analyze it and publish using it if they find something novel. It’s really a resource for the entire academic community.”
St. Jude Cloud accelerates discovery through intuitive summary visuals
Within St. Jude Cloud, multiple ways exist to look at genes, chromosomes, epigenetics, mRNA and protein data. GenomePaint presents the user with summary visuals of the genetics and epigenetics of a desired group of patient samples, such as those of a particular subtype. ProteinPaint illustrates sequence features proteins. Users can use it to examine the domains of genes, known isoforms of a given gene, hotspot mutations for single nucleotide variations (SNVs), insertions and deletions (Indels) and structural variations (SVs) in both pediatric and adult cancers, and RNA-seq expression of a given protein in different tumor types.
These flagship tools were created by the team of Xin Zhou, Ph.D., St. Jude Department of Computational Biology. The representation of sequence variation in Binary Alignment Map (BAM) files in ProteinPaint is so user-friendly and convenient it was recently published in Bioinformatics. It will also soon be incorporated into the National Cancer Institute’s Genomic Data Commons (GDC) portal, the central sequence repository for cancer research.
When scientists want to show a specific tissue or cancer type’s mutations and protein expression information, they can create an interconnected Oncoprint visualization with these resources. Additionally, they portray a landscape, often for a given subtype. For example, the Oncoprints can summarize what is “expected” for that given subtype. Researchers are asked to curate their Oncoprints; this ensures a user isn’t looking at noise or false positives but the most meaningful summary data.
“Oncoprints are a great tool to get an overview of genes across several samples,” Karlstrom said. “For example, if there are known or novel fusions. It can also help you recognize if a new case study or newly published mutation is prevalent in the disease you study. It’s easy to go in and take a look to ask, ‘Do we have any of those fusions in our data?’”
While the Oncoprint is a useful high-level summary of mutations, researchers may also be interested in how epigenetics regulate or dysregulate genes. St. Jude Cloud leverages an epigenetics viewer in GenomePaint, built by Xhou, that scientists can look at to gauge the local context of epigenetic markers.
“The epigenetic visualizations give a very visual guide,” Karlstrom said. “You can see the marks. If you have a distinct region you want to look at, you can easily see what state it is.”
Within the platform, researchers can look at extensive overviews of a region of DNA or RNA or zoom in on a specific site. Common variants are demarcated within a chosen tissue or cancer subtype. Not only does this show researchers the development stages, but it also includes tissue-type specific data. The RNAseq data contains thousands of samples. In addition, St. Jude Cloud houses a small number of single-cell RNAseq (scRNAseq) visualizations.
“The Cloud hosts some awesome single-cell RNAseq visualizations,” Karlstrom said. “For example, there are single-cell and single-nucleus RNAseq for rhabdomyosarcoma and neuroblastoma samples. Within the Cloud, the users can change the dynamics of these scRNAseq visualizations to look at them differently. Scientists can investigate the visualization the way they want to and get answers. It’s a huge contribution to academic research, making this data accessible in these visualizations.”
St. Jude Cloud made interoperability an essential part of the system to aid the ease of use. A user is not restricted to one point of view, such as genomics, epigenetics, RNA or protein level data. In fact, they are not restricted to one particular cancer subtype or model. Instead, the platform links these details together in a logical fashion.
St. Jude Cloud facilitates data sharing and novel research
For simplicity, researchers can access some of these high-level visualizations without needing to go through the entire process of a data access agreement. They can then perform a pilot, looking through the visualized summary information, before pursuing the necessary steps to gain full data access.
If scientists want to use raw data, the team created St. Jude Cloud to accelerate discovery by removing barriers to access. One intrinsic barrier to using large datasets is the difficulty of accumulating and processing them. Even simple tasks, such as finding physical storage and downloading these files can be an issue. Scientists would have to take samples and spend a large sum on sequencing them all, as making the sequences publicly available is required for publication.
If scientists have access to already sequenced samples, it removes several steps. Instead, they can save resources to apply to another assay or experiment other than sequencing. St. Jude Cloud adds even more value by providing both raw files and visualizations. Scientists can access these raw files or the processed data as visualizations, whichever is more advantageous.
Karlstrom works for the St. Jude Childhood Solid Tumor Network (CSTN), a resource where academic researchers can access xenografts and associated data from over 22 solid tumors diagnosed without an obligation to collaborate. Since its inception, the CSTN has provided xenografts to hundreds of labs worldwide. St. Jude Cloud enabled more academic researchers to unlock xenograft research to stimulate basic research and speed up translation to the clinic.
“Before we built the data portal and the Cloud had all these visualizations, sharing associated data generated but not published was not as easy,” Karlstrom explained. “But now the data is available, and we’ve already sequenced the tumors they can request through CSTN.” CSTN also has samples that have not yet been published, but all data and samples are still shared.
This sequencing data is also paired with other valuable information. Researchers can view a number of other visualizations of data collected on any given sample.
“The Cloud includes other characteristics, such as immunohistochemistry and electron microscopy images and previous testing done on the samples. Therefore, scientists don’t have to expend the resources in their lab to do these baseline analyses,” Karlstrom explained.
Scientists can make significant discoveries using the whole genome, whole exome and whole transcriptome data stored and visualized on St. Jude Cloud, especially when paired with other information, such as histology. But St. Jude went even further to accelerate discovery by fostering more collaborations.
Sharing visualizations for collaboration
St. Jude Cloud includes a Visualization Community (VisCom) to facilitate collaboration. For example, VisCom includes groups based on specific topics, such as the audacious goals initiative — retina (AGI Retina), Neuro-Oncology, Sickle Cell or Cancer Survivorship Community. Users can share their visualizations publicly after publication or privately between collaborators before publication.
“The newly released version of PeCan is very exciting because any user can go there and look at their favorite gene,” Karlstrom said. “And then, they can reach out to potential or current collaborators, then show what they are thinking with the visualization.”
As the program manager for CSTN, Karlstrom often receives requests from interested researchers looking for xenografts from St. Jude. St. Jude Cloud provides an accessible resource to begin a conversation.
“People reach out to CSTN,” Karlstrom said. “They either have a gene they know is involved in some system they’re looking at, and they want to know, ‘How prevalent is the gene mutation? Is it linked to something else? Is there a fusion? Is it susceptible to specific kinds of drugs? Is there any indication that patients who are a certain age have it?’ And now it’s possible for people to delve into the data and visualizations themselves.”
That initial conversation can guide scientists to a specific and important research question.
“To me, the biggest gain here is that you can go in and look, and we have these beautiful visualizations for individual diseases. You can narrow your research focus to the most interesting questions,” Karlstrom explained.
St. Jude Cloud helps user through support chat
Each visualization from St. Jude Cloud contains a large amount of information. Starting a visualization is guided; for example, a user can copy the interfaces that use actual sample data, then customize them. Or, if the user is an expert, they can use the Visual Editor (VisEditor) feature directly if they want to code the visualization to meet their exact specification. Even with these intuitive visualizations, with so many features, datasets and capabilities, Karlstrom sometimes needs to ask for help.
The St. Jude Cloud team staffs a support chat function to help answer any user’s questions. Instead of an automated response, a real cloud expert is a simple chat question away.
“It’s great, that little button on the right,” Karlstrom said. “You write your question there, and then you get a response and can easily find out what you need. They are very attentive, and the response time is very quick. In my experience, they have responded in a very, very short timeframe, often within minutes to an hour.”
If a scientist needs help, the St. Jude Cloud team is ready to be a resource to problem-solve, troubleshoot and otherwise support the researcher so they can maximize their time pursuing their studies and minimize the time spent experiencing technical difficulties. To that end, the team also hosts a Help Guide and tutorials so scientists can learn how to get the most out of the platform on their own time.
St. Jude Cloud visualizations are pushing science forward
Each feature of St. Jude Cloud visualization applications, from data presentation to VisCom to the support chat, was designed to facilitate research. The team created the resource to be useful during data discovery, analysis and preparation for publication. The scientific community already uses these features to publish peer-reviewed articles with powerful data visualizations. For example, using the applications, 38 papers from 20 VisCom teams have been published in journals from Cancer Discovery to Nature. VisCom contains over 79 curated visualizations, both published and unpublished, a user can explore to facilitate their research and lead to their own publications.
In St. Jude Cloud, the scientific community has a powerful tool to visualize data and answer the most important scientific questions.